Sequence Confirmation of Oligonucleotides through Collision Induced Dissociation Tandem Mass Spectrometry
Matthew Crawford, MS
In the realm of molecular biology, the elucidation of DNA and RNA sequences is essential to furthering
research and maintaining high therapeutic quality. Oligonucleotides, short nucleic acids, play a crucial
role in various biological processes and are extensively employed in research, diagnostics, and thera-
peutics. However, ensuring the reproducibility of synthesized oligonucleotides is fundamental for their
intended applications. This is where advanced analytical techniques like Collision Induced Dissociation
Tandem Mass Spectrometry (CID-MS/MS) step in, offering a powerful means of sequence confirmation.
Unveiling the Technique: Collision Induced Dissociation Tandem Mass Spectrometry (CID-MS/MS)
CID-MS/MS is a tandem mass spectrometry technique known for its ability to provide detailed structur-
al information about molecules. In the context of oligonucleotide analysis, CID-MS/MS offers insights
into the sequence, modifications, and purity of synthesized oligonucleotides. Let’s look at the mecha-
nism behind this technique:
- Ionization: The process begins with the ionization of oligonucleotide molecules, typically through
electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI). This step gen-
erates ions suitable for mass spectrometric analysis. - Selection and Activation: Once ionized, the oligonucleotide ions are selected based on their
mass-to-charge ratio (m/z) and subjected to collision-induced dissociation (CID). During CID, the
selected ions are collided with inert gas molecules (e.g., helium or nitrogen), inducing fragmenta-
tion. - Fragmentation Analysis: The collision energy applied during CID causes the cleavage of phos-
phodiester bonds within the oligonucleotide backbone, resulting in the generation of sequence-
specific fragment ions. These fragment ions retain information about the nucleotide sequence,
allowing for sequence elucidation. - Mass Analysis: The resulting fragment ions are then analyzed based on their mass-to-charge
ratios, enabling the determination of the oligonucleotide sequence. By comparing the observed
fragment ions with theoretical fragment patterns, the sequence of the oligonucleotide can be con-
firmed.
Advantages of CID-MS/MS for Oligonucleotide Analysis
CID-MS/MS offers several advantages for oligonucleotide analysis:
- High Sensitivity: CID-MS/MS can detect oligonucleotides at low concentrations, making it suitable
for analyzing minute quantities of synthesized sequences. - Sequence Specificity: The fragmentation patterns obtained through CID-MS/MS provide sequence-
specific information, enabling accurate sequence confirmation not available through simple intact
mass confirmation. - Speed and Efficiency: The technique offers rapid analysis, allowing for the high-throughput charac-
terization of oligonucleotide samples. - Compatibility with Modifications: CID-MS/MS can also characterize oligonucleotides with various
modifications, such as phosphorylation, methylation, base modifications, or base-linkage
(phosphorothioate) modifications, enhancing its versatility.
Applications of CID-MS/MS in Oligonucleotide Research
The application of CID-MS/MS in oligonucleotide research is multifaceted:
- Quality Control: CID-MS/MS is employed for quality control purposes to ensure the accuracy and
integrity of synthesized oligonucleotides. - Structural Elucidation: The technique facilitates the elucidation of complex oligonucleotide struc-
tures, including sequence variants and modifications. - Biomedical Research: CID-MS/MS plays a vital role in biomedical research, including the study of
gene expression, RNA editing, and the development of targeted nucleic acid-based therapeutics.
“Poly-T” Ladder Sample Set
In this example set (“Poly-T” Ladder Sample) we did work with what we called T10.00 (TTTTTTTTTT) and
T10.02 (TTTT^TT^TTTT, where ^ indicates phosphorothioate linkages). T10.00 is a normal DNA oligo
without any modifications. T10.02 is a DNA oligo modified with 2xphosphorothioate modifications, a
common modification used to stabilize RNA and DNA structures for therapeutics. The phosphorothio-
ate modification inhibits endo- and exonuclease activity, increasing drug half-life in the body.
T10.00/T10.02 were both analyzed by MSMS, which selects a precursor and then fragments it as it goes
through the QToF collision cell. Based on the spectra and the software predictor capabilities we can
confirm the fragments.
The data were analyzed in Agilent Bioconfirm using the Sequence Confirmation workflow.
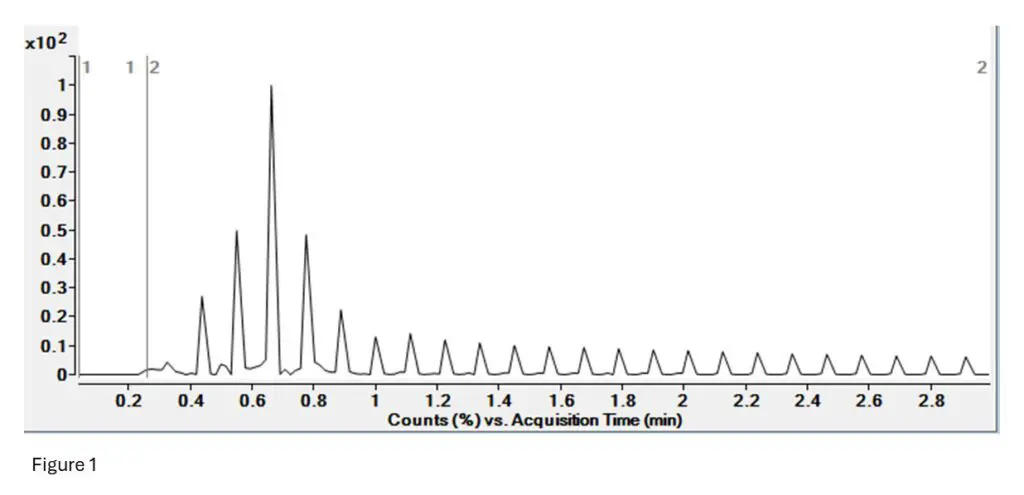
Figure 1: This is the TIC (Total Ion Current) for the T10.00 run. There is no LC separation as this was
done by FIA (Flow Infusion Analysis). The spikes indicate selection of a precursor, while the valleys are
the fragmentation of that precursor into its structural pieces.
Essentially, we see a large signal when the selection starts, and then see low signal during selection as
all other ions that are not the one we are looking at are excluded (pattern repeats every few seconds).
This is the same for peptide mapping by MSMS as well, or only DDA-MSMS workflow.
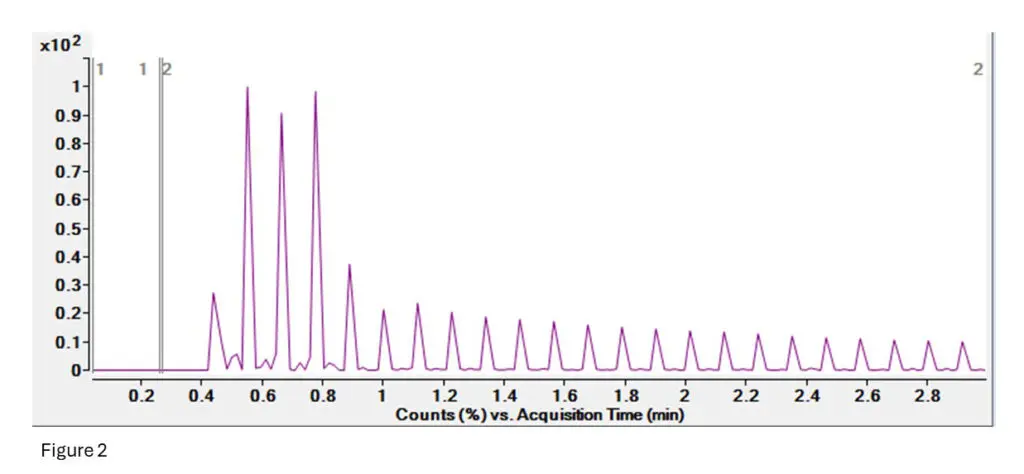
Figure 2: This is the TIC (Total Ion Current) for the T10.02 run.
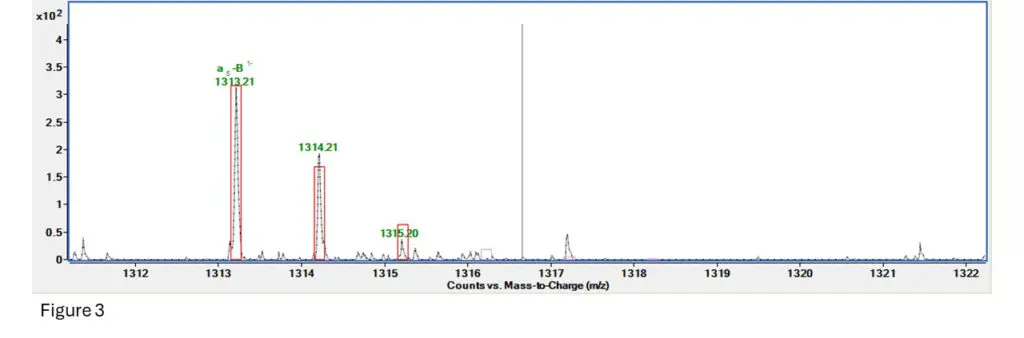
Figure 3: This is the fragment spectrum for a section of T10.00, specifically TpTpTpT (4 bases and 3
phosphodiesters, while the 4th base has lost its phosphate group).
The software predicts fragmentation based on an algorithm and generates ranges for how much and
how far apart the fragment ion isotope clusters should be. It then creates a box around where it expects ions to be and gives the fragment a score based on how many of those ions are inside the range
provided by the box.
Here we can see that the first ion is right inside it, the second one is a bit more abundant than expected
but still close enough, and the third is a bit less abundant than expected but is present.
Fragment confirmed!!
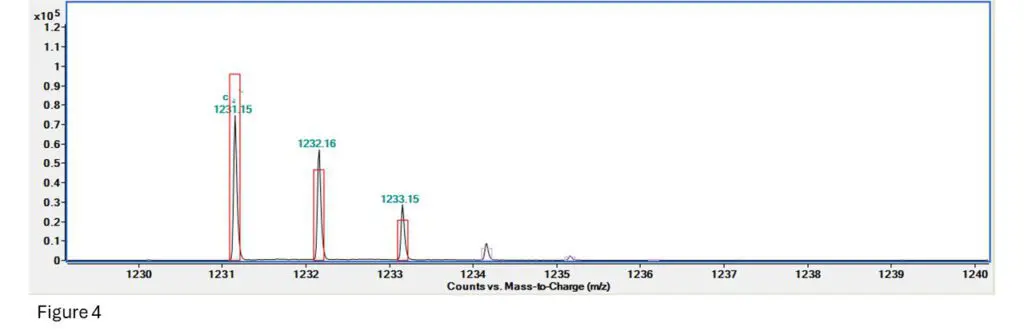
Figure 4: This is the fragment spectrum for a section of T10.02, specifically TpTpTpTps (so 4 bases and 3
phosphodiesters, plus 1 phosphorothioate.
Here we can see that the signal for the first ion is a bit low, the second one is a bit more abundant than
expected but still close, and the 3rd, 4th and 5th are all present but a little extra abundant.
Fragment confirmed!!
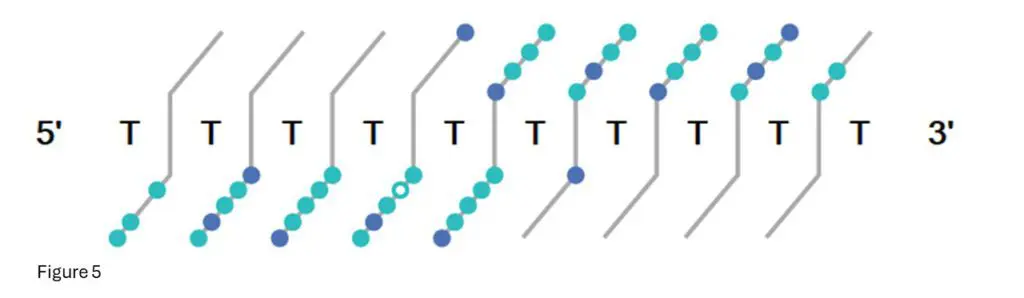
Figure 5: This is the fragment confirmation ladder from Bioconfirm for T10.00.
Each light-blue dot indicates a confirmed fragment isotope cluster from that base position, while dark
blue dots indicate multiple fragments confirmed.
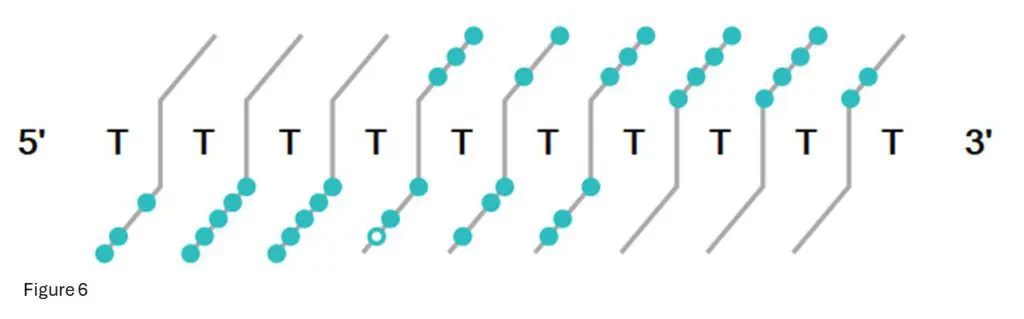
Figure 6: This is the fragment confirmation ladder from Bioconfirm for T10.02
Each light-blue dot indicates a confirmed fragment at that base.
Conclusion
Collision Induced Dissociation Tandem Mass Spectrometry (CID-MS/MS) serves as a cornerstone tech-
nique for the sequence confirmation of oligonucleotides. Its ability to provide high sensitivity, sequence
specificity, and compatibility with modifications makes it indispensable in the field of molecular biology
and biotechnology. As research advances, CID-MS/MS is poised to continue unraveling the complexities
of nucleic acid sequences, fostering innovation and discovery in diverse scientific domains.
