Understanding the Impact of Non-Volatile Salts in Mass Spectrometry Analysis
Matthew Crawford, MS
Mass spectrometry (MS) has revolutionized the field of proteomics and genomics, enabling researchers to
unravel the complex structures and functions of proteins and nucleic acids with unprecedented precision.
However, despite its power, MS analysis can be influenced by various factors, including the presence of non-volatile salts in samples. In this blog post, we’ll delve into the effects of non-volatile salts on MS analysis of oligonucleotides and explore strategies to mitigate their impact.
Non-volatile salts are commonly used in oligonucleotide sample preparation and purification processes.
These salts, such as sodium chloride (NaCl), play crucial roles in oligo solubilization, precipitation, and
chromatographic separation. However, their presence in MS samples can lead to several challenges that affect the accuracy and sensitivity of nucleic acid analysis.
One of the primary concerns with non-volatile salts in MS analysis is ion suppression. When samples
containing high concentrations of salts are introduced into the MS system, the ionization efficiency of analytes may decrease due to competition for ionization with salt ions. As a result, low-abundance analyte ions may be masked by the intense signals from salt ions, leading to reduced sensitivity and compromised detection limits.
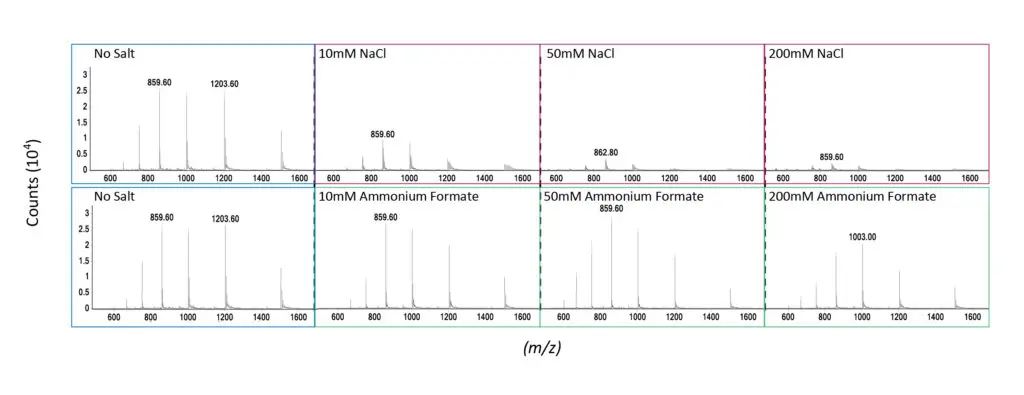
salt (sodium chloride) at various concentrations when analyzing a 20mer oligonucleotide. As can be seen,
the signal intensity for the analyte ions drops considerably for the non-volatile salt as salt concentration
increases, while volatile salts have a much smaller effect.
Moreover, non-volatile salts can also cause signal suppression or enhancement/adduction effects in MS
detection. These effects arise from the formation of adducts between oligo ions and salt ions during the
ionization process. For example, anionic oligos may interact with cationic salt ions (such as Tris), leading to signal enhancement. These adduct formations can distort the true abundance of protein ions and complicate data interpretation.
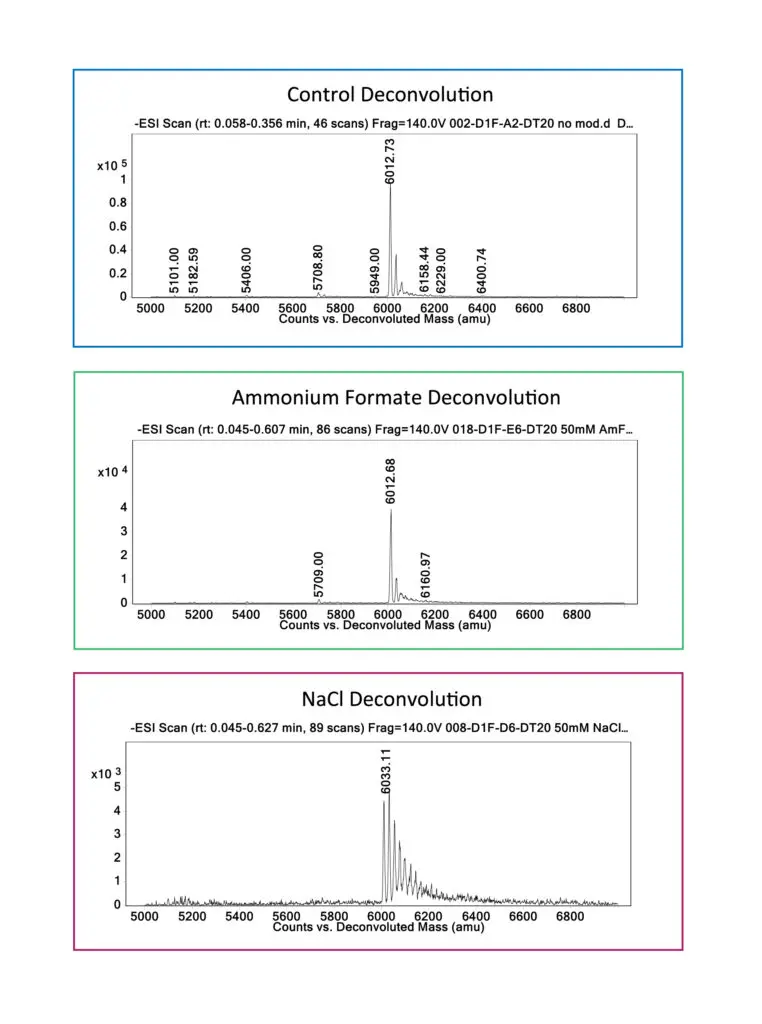
salts such as ammonium formate dissociate during ionization, leading to a single major intact mass. By
contrast, sodium ions from sodium chloride remain attached to the oligo, generating a series of peaks
resulting from singly adducted and multiply adducted species.
To address the challenges posed by non-volatile salts in MS analysis, several strategies can be employed:
1. Salt removal techniques: Various methods, such as dialysis, ultrafiltration, solid-phase extraction, or liquid-liquid extraction, can be used to remove non-volatile salts from protein samples before MS analysis. These techniques help minimize ion suppression and improve the sensitivity of analyte detection.
2. Optimization of sample preparation: Careful optimization of sample preparation protocols, including
salt concentration, pH, and buffer composition, can help reduce the impact of non-volatile salts on MS
analysis.
3. Use of volatile buffers: Utilizing volatile buffers, such as ammonium acetate or ammonium formate, for
oligo solubilization and chromatographic separation can prevent the introduction of non-volatile salts into the MS system. Volatile buffers evaporate readily during the ionization process, minimizing interference with protein ionization (and reducing adduct formation) and improving overall MS performance.
4. Online desalting techniques: Incorporating online desalting techniques, such as liquid chromatography
coupled with MS (LC-MS) or capillary electrophoresis-MS (CE-MS), can effectively remove non-volatile
salts before analyte ionization. These techniques enable simultaneous sample cleanup and analysis,
reducing sample preparation time and improving data quality – and this is a Rilas specialty!
In conclusion, non-volatile salts can significantly impact the accuracy and sensitivity of MS analysis of all
analytes by causing ion suppression and signal distortion effects. However, by employing appropriate sample preparation strategies and optimizing experimental conditions, researchers can mitigate these challenges and enhance the reliability of molecule identification and quantification in MS-based -omics studies. Understanding the effects of non-volatile salts is essential for advancing our knowledge of complex biological systems and uncovering novel insights into biomolecule structure and function.
